understand axis in matplotlib
axis is a subarea within figure() in matplotlib
figure()
plt.axes([0.3, 0.5, 0.4, 0.2])
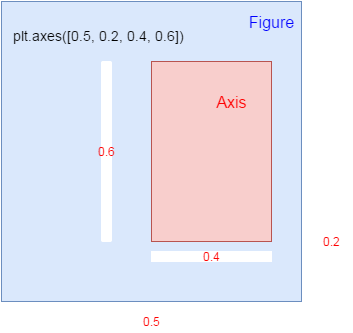
the figure area size is 1, 1 as base
|
In [2]:
import matplotlib.pyplot as plt
import numpy as np
In [6]:
physical_sciences=[ 13.8, 14.9, 14.8, 16.5, 18.2, 19.1, 20. , 21.3, 22.5,
23.7, 24.6, 25.7, 27.3, 27.6, 28. , 27.5, 28.4, 30.4,
29.7, 31.3, 31.6, 32.6, 32.6, 33.6, 34.8, 35.9, 37.3,
38.3, 39.7, 40.2, 41. , 42.2, 41.1, 41.7, 42.1, 41.6,
40.8, 40.7, 40.7, 40.7, 40.2, 40.1];
computer_science=[ 13.6, 13.6, 14.9, 16.4, 18.9, 19.8, 23.9, 25.7, 28.1,
30.2, 32.5, 34.8, 36.3, 37.1, 36.8, 35.7, 34.7, 32.4,
30.8, 29.9, 29.4, 28.7, 28.2, 28.5, 28.5, 27.5, 27.1,
26.8, 27. , 28.1, 27.7, 27.6, 27. , 25.1, 22.2, 20.6,
18.6, 17.6, 17.8, 18.1, 17.6, 18.2]
year=[1970, 1971, 1972, 1973, 1974, 1975, 1976, 1977, 1978, 1979, 1980,
1981, 1982, 1983, 1984, 1985, 1986, 1987, 1988, 1989, 1990, 1991,
1992, 1993, 1994, 1995, 1996, 1997, 1998, 1999, 2000, 2001, 2002,
2003, 2004, 2005, 2006, 2007, 2008, 2009, 2010, 2011]
two methods to specify many distinct graphs¶In [10]:
# Plot in blue the % of degrees awarded to women in the Physical Sciences
plt.plot(year, physical_sciences, color='blue')
# Plot in red the % of degrees awarded to women in Computer Science
plt.plot(year, computer_science, color='red')
# Display the plot
plt.show()
In [8]:
# Create plot axes for the first line plot
plt.axes([0.05,0.05,0.425,0.9])
# Plot in blue the % of degrees awarded to women in the Physical Sciences
plt.plot(year,physical_sciences, color='blue')
# Create plot axes for the second line plot
plt.axes([.525,0.05,0.425,0.9])
# Plot in red the % of degrees awarded to women in Computer Science
plt.plot(year,computer_science, color='red')
# Display the plot
plt.show()
In [9]:
# Create a figure with 1x2 subplot and make the left subplot active
plt.subplot(1,2,1)
# Plot in blue the % of degrees awarded to women in the Physical Sciences
plt.plot(year, physical_sciences, color='blue')
plt.title('Physical Sciences')
# Make the right subplot active in the current 1x2 subplot grid
plt.subplot(1,2,2)
# Plot in red the % of degrees awarded to women in Computer Science
plt.plot(year, computer_science, color='red')
plt.title('Computer Science')
# Use plt.tight_layout() to improve the spacing between subplots
plt.tight_layout()
plt.show()
In [ ]:
|